Artml for pneumonia detection
Pneumonia is a form of a respiratory infection that affects the lungs and it is the single largest infectious cause of death in children worldwide. According to the World health organization, Pneumonia killed 920136 children under the age of 5 in 2015 alone, accounting for 16% of all deaths of children under five years old. When an individual has pneumonia, the parts of the lungs are filled with pus and fluid, which makes breathing painful and limits oxygen intake.
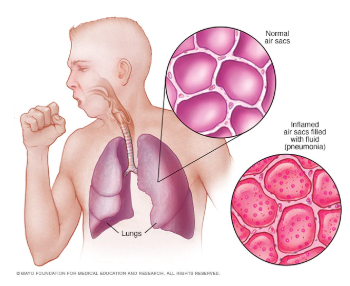
Detecting infection in early stages is crucial for curing pneumonia. Chest X-rays are currently the best available method for detecting this. However, using chest X-rays is a challenging task since it requires expert radiologists for making the decision.Also, statistics show that Pneumonia is most prevalent in South Asia and sub-Saharan Africa where there are only a few experts to classify chest X-ray for abnormalities. Hence there is a lot of research in this area for building models that can automatically detect pneumonia from chest X-rays. State of the art models like CheXNet developed by Stanford ML group is making detections at a level exceeding practicing radiologists. One significant problem with the current solution is that it doesn’t leverage the vast amount of labeled data which is generated every day. Using this data could make models more robust and effective for pneumonia detection. Current work explores ARTML methodology for building models that are scalable and giving the power of continual learning.
Data Preparation:
Two datasets Chest Xray images and RSNA Pneumonia data from Kaggle are used for building the ARTML model. Dataset-1 has 5218 training images and dataset 2 has 26684 images respectively. Testing data is considered from data 1 which has 604 images. Images are in DICOM format - which is the standard file format for medical information. All these images are imported in python environment and converted into JPG format with 150*150-pixel size.
Reason for using two datasets is to gather varied images for training so as to make the model more robust. Also, generally building Deep models using large data is a challenging task - we used this large corpus of data here to prove how ARTML scalability power could make this task easy even with limited computational power.
Data Exploration:
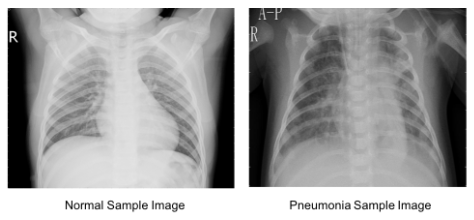
Below images show the Normal lungs and the one affected with pneumonia. In the affected Lung image, we can observe Opacity due to the haziness. But in many cases, it is difficult to differentiate between the opacity caused by pneumonia or other temporary reasons just by observing the Chest X Rays. Below figure clearly shows such challenging cases which shows the need for expert-level decisions for predicting the actual cause (whether pneumonia or not).
Modeling:
Initially, convolutional network is built using the Chest Xray images and [RSNA Pneumonia data] dataset which has around 5200 training images. After building the CNN network ARTML QDA and Naïve Bayesian models are trained on the extracted features. As discussed earlier since the idea of using ARTML is also to address the scalability issues, all the remaining image data (25000 images ≈5GB) are incrementally updated in the model using ARTML inbuilt functions. No cloud or GPU power is used for training the image data, all the models are trained on a standard laptop with 8GB RAM. This clearly proves the scalability power of ARTML models. Look into the Jupyter notebook for detailed code.
Evaluation:
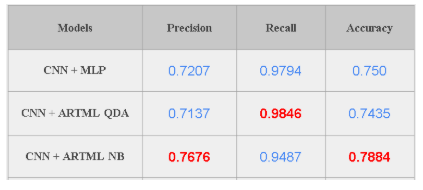
Model is tested on a dataset which has 624 images. When the ARTML model is used for predictions we get 79% accuracy for Naive bayes classifier and 74% accuracy for QDA. But accuracy is not a very good measure for medical image classification. Since in this scenario, True positive cases are very important (We want to correctly identify all of the Pneumonia cases) Recall could be a good metric for evaluating the model performance. Our final model has a Recall of 98.46% on the test images implying that the model is able to predict 98% of all pneumonia cases correctly. On the other hand, we observe that precision is low for the model - 74-78%. Since Precision and Recall follows a trade-off, and we need to find a point where recall, as well as precision, are in the acceptable levels.
Ensembles: To further improve model performance different CNN feature extractors could also be built using the initial training data and use all the extracted features for building different ARTML models. Final prediction could be decided based on either the majority of the predictions or using the average of the predicted probabilities on the testing data.
Deployment:
Power of ARTML models can be truly identified during the deployment. Since in this particular case labeled images are generated every day by the Radiologists, we can leverage this data to update the models real time in a computationally cheaper way. Every single scanned X-Ray image which is wrongly classified by the model could be used to update the model in real time. The main reason for this continuous retraining of the model is to avoid any further misclassifications if similar images are encountered in the future. Traditional Deep neural networks lack this flexibility making the ARTML approach more powerful. Also as ARTML supports decremental learning, if any wrongly labeled X-Ray data is used in retraining the model - this effect could be easily reverted using the models forgetting behavior.
Trained ARTML model is updated using the Test data in an incremental way to verify the above hypothesis (if the same retained images are used for testing whether the model can correctly identify them or not). Results show that Recall and precision significantly improved.
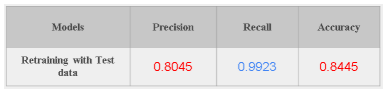
Above results clearly show that by updating the model continuously it will help to reduce the model bias over time. Overall we can observe that ARTML is on par with the existing state of the art deepnets in its performance, additionally giving flexibilities to learn/forget in real time just by using limited computational resources.